Abstract
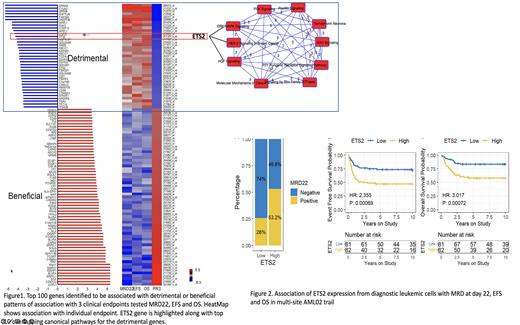
Pathogenicity of AML is characterized by many chromosomal and gene alterations, which summarize substantial differences in response to therapy and survival of patients following chemotherapy. More in-depth understanding of the molecular and genomic landscape of AML is paramount to improving the therapeutic outcome and prospecting new targets to help combat the battle against AML. Genome-wide transcriptomics-based approaches have shown promising results in identifying signatures associated with poor outcomes. Our group has previously developed and applied an innovative PROMISE (PRojection Onto the Most Interesting Statistical Evidence) statistical method to enable simultaneous and directional evaluation of multiple clinical endpoints against an omic variable (1). In our previous investigations, we used a cohort of 42 patients from the multi-site AML97 clinical trial and a cohort of 46 patients from multi-site AML02 (NCT00136084). to identify genes associated with the biologically meaningful and therapeutically relevant pattern of association with multiple clinical and pharmacologic endpoints (2). Given the small sample size investigated in previous report, we now extended PROMISE analysis, we extended PROMISE analysis (unadjusted and risk-adjusted analysis) using three clinical endpoints (Minimal Residual Disease at day 22 (MRD22), Event Free Survival (EFS), and overall survival (OS)) in 163 patients treated on multi-site AML02 clinical trial (NCT00136084). At the PROMISE p-value threshold of <0.001, 177 probes mapping to 153 unique genes were identified, of these, 59 were associated with detrimental pattern (MRD positivity and inferior EFS and OS) of outcome association, and 94 were associated with beneficial pattern (MRD negativity and better EFS and OS) of association with clinical outcome. Fig 1 shows the bar-plot and heat-map for the top 100 genes identified in the integrated PROMISE analysis. As expected, DNMT3B, GPR56, SPINK2, and FAM30A, part of our recently reported leukemic stem cell scores, were among the top detrimental hits (MRD1, EFS, OS, and PROMISE, all p values <0.0002). Pathway analysis of 59 genes associated with detrimental outcomes using Reactome, Ingenuity pathway analysis, and other pathway analysis tools identified pathways enriched with the genes significantly associated with outcome. We want to highlight a few detrimental genes worth pursuing as prognostic biomarkers or for novel drug development. ETS2 plays a role in 'Oncogene Induced Senescence,' which is triggered by high levels of RAS/RAF/MAPK signalling. ETS2 is also involved in HER2 signalling in breast cancer and HGF signaling (Fig 1). In our dataset, ETS2 was significantly associated with higher MRD22 positivity and inferior EFS and OS (all p<0.002), Fig 2. ETS2 was further expressed at a higher level in AML cells than normal (p<0.05). Though the role of ETS2 in adult AML has been recently reported, our results in paediatric AML further support it as a potential therapeutic target for pursuing drug development. Among the significant detrimental genes with potential actionable drugs included drug transporter ABCC1, a glycoprotein 2A, a transmembrane receptor ITGA2B and YES1, a protooncogene belonging to SRC tyrosine kinase superfamily. Overall, our integrated analysis to identify genes that are predictive of a therapeutically meaningful patterns of association with multiple clinical outcome endpoints identified markers of significant prognostic and therapeutic relevance; ongoing studies are focused on PROMISE validation analysis in an independent cohort of patients from AML08 clinical trial.
References: 1. Pounds S, Cheng C, Cao X, et al: PROMISE: a tool to identify genomic features with a specific biologically interesting pattern of associations with multiple endpoint variables. Bioinformatics 25:2013-9, 2009
2. Lamba JK, Crews KR, Pounds SB, et al: Identification of predictive markers of cytarabine response in AML by integrative analysis of gene-expression profiles with multiple phenotypes. Pharmacogenomics 12:327-39, 2011
Acknowledgements: National Institute of Health (NIH) grants R01-CA132946 (Lamba and Pounds), University of Florida Opportunity Seed Grant (Lamba), American Lebanese Syrian Associated Charities (ALSAC). We thank Drs. Campana, Coustan-Smith for MRD data, Dr. James Downing for Gene-expression and Dr. Susana Raimondi for the cytogenetics data.
Disclosures
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.